|
|
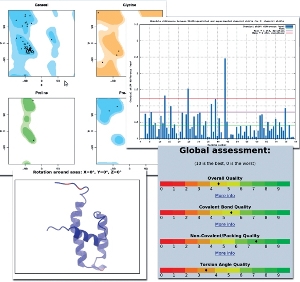
PROSESS (Protein Structure Evaluation Suite & Server) is a
web server designed to evaluate and validate protein structures solved by either X-ray crystallography or NMR spectroscopy.
PROSESS integrates a variety of previously developed, well-known and thoroughly tested methods to evaluate both global and
residue-specific: 1) covalent and geometric quality; 2) non-bonded/packing quality; 3) torsion angle quality;
4) chemical shift quality and 5) NOE quality.
In particular, PROSESS uses VADAR
for coordinate, packing, H-bond, secondary structure and geometric analysis, GeNMR
for calculating folding, threading and solvent energetics, ShiftX
for calculating chemical shift correlations, RCI for
correlating structure mobility to chemical shift and Preditor
for calculating torsion angle-chemical shifts agreement. PROSESS also incorporates several other programs including
MolProbity
to assess atomic clashes and His/Asn flips, XPLOR-NIH to identify and quantify NOE restraint violations and
NAMD to assess structure energetics.
PROSESS produces detailed tables, explanations, structural images and graphs that summarize the results and compare them to values
observed in high-quality or high-resolution protein structures. Using a simplified red-amber-green coloring scheme PROSESS also
alerts users about both general and residue-specific structural problems. PROSESS is intended to serve as a tool that can be used by
structure biologists as well as database curators to assess and validate newly determined protein structures.
Reference:
PROSESS: a protein structure evaluation suite and server;
Berjanskii M, Liang Y, Zhou J, Tang P, Stothard P, Zhou Y, Cruz J, Macdonell C, Lin G, Lu P, Wishart DS.;
Nucleic Acids Res. Webserver Edition; 2010.
|
|
| 1) Enter your email address (Optional)
|
| 2) Upload coordinates of a 3D protein model in the PDB format (Required)
|
| 2) Upload primary sequence in FASTA format if the 3D model has missing residues (Optional) |
| 3) Upload distance restraints with residue numbers matching the 3D model (Optional)
|
| 4) Upload chemical shifts in BMRB NMR-STAR 2.1 or NEF formats with residue numbers matching the 3D model (Optional) |
5) Press the submit button. The results will be displayed on the web in 5-30 min depending on your model.
If you enter email address, you will also receive an email with links to results.
|
6) Print PROSESS results as soon as you receive email notification that your PROSESS job is finished.
PROSESS job files are stored on the server for 3 days.
Important!!!
- Your PDB file, NOE file, and chemical shift file must have identical residue numbering.
- Residue numbering for sub-units of dimers, tetramers, and other multimers must not overlap.
- Chemichal shifts of sub-units must be combined into a single list in NMR-Star 2.1 format
with combined primary sequence entries.
- If you submit an ensemble, the minimal size of an ensemble should be 3 models.
|

